

Initiatives & Research
BBJ began its fifth phase of operations in a fiscal year 2023. In recent years, genomic and omics analysis methods have been rapidly developed with advances in next-generation sequencing and other analytical technologies. As a result, Genomic and Omics analysis data are widely used for genome research including biomarker discovery research and other researches aimed at developing new clinical and therapeutic methods, Against this backdrop, BBJ is working for genomic and omics analysis of biological samples collected from the patients for further enhancing its data base for genomic medical research with the aim of implementing genomic medicine.
BBJ is enhancing a multi-omics data infrastructure focused on liver diseases, autoimmune and allergic diseases, dementia, cardiovascular diseases, and infectious diseases etc.
To build a multi-omics data infrastructure and promotion of research utilization of data
NASH/NAFLD, autoimmune and allergic diseases, dementia, cardiovascular diseases, and infectious diseases.
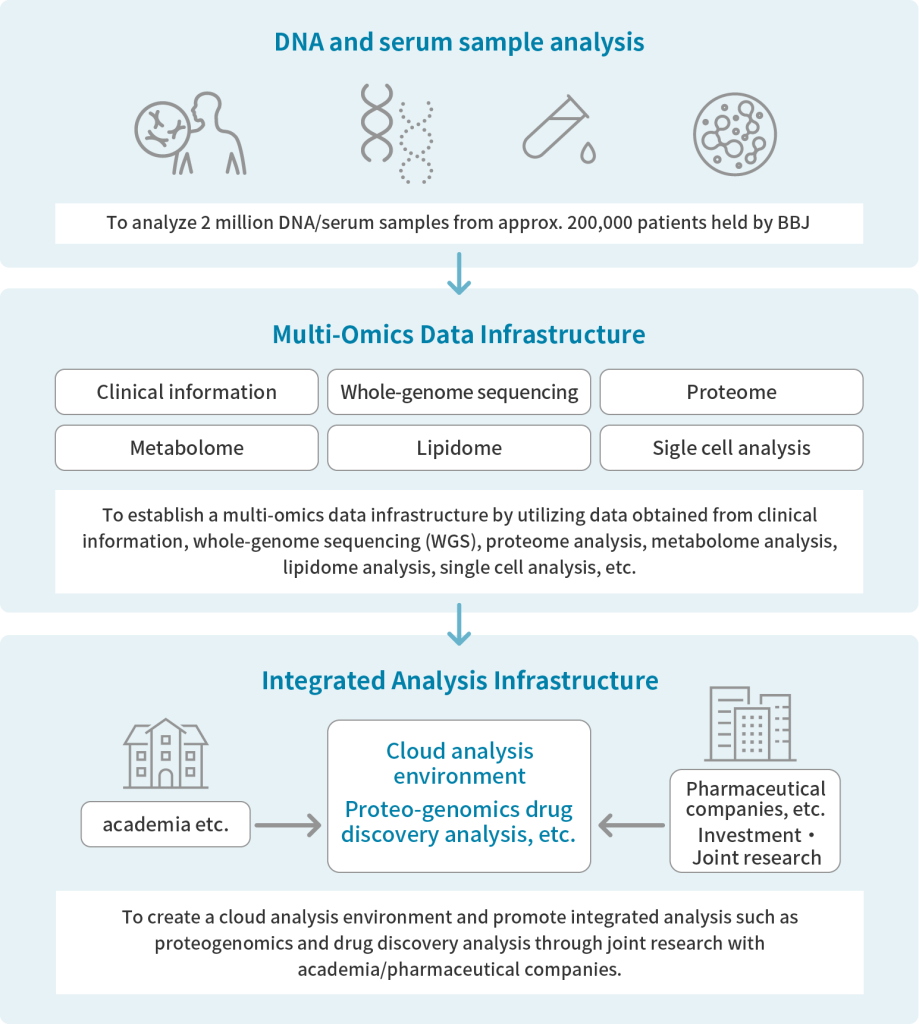
As of November 2023, we have completed all SNP array analyses of samples from the 1st cohort (200,000 patients) and the 2nd cohort (67,000 patients) and have made the data available. These data have been widely used in genome-wide association analysis (GWAS) studies to detect DNA polymorphisms associated with the diseases under study. We have also completed whole genome sequencing (WGS) of 4,800 patients in the 1st cohort and 6,000 individuals in the 2nd cohort and have made the data publicly available at BBJ and in public databases.
For omics analysis, we have completed metabolomic analysis of 4,000 samples from the 1st cohort and are preparing to release data for analysis of an additional 60,000 samples. Metabolomics, the lowest level of central dogma, is important in defining the relationship between both genetic and environmental factors and disease. Comprehensive metabolome analysis is also expected to be used in the search for molecular biomarkers, such as narrowing down metabolites that fluctuate specifically for a particular disease. Proteome data for 3,000 participatns from the 1st cohort has been made public since June 2025.
Please refer to the information on the 1st and 2nd cohort at the following page.
For all the publicly available Genomic and Omics data, please refer to the following webpage.