

For Researchers
In addition to the BBJ’s own database, we also make genome and omics analysis data available from public databases such as the NBDC human database and AMED Genome Group Sharing Database (AGD). This enables BBJ to offer extensive genome and omics analysis data, empowering researchers to seamlessly integrate BBJ data into their studies and enhance data utilization across scientific communities.
*Starting Apr. 1, 2024, the Database Center for Life Science (DBCLS) operates the NBDC human database. Accordingly, URLs of the web site and the links related to the site have been changed.
BBJ manages and stores the data listed below in the BBJ database. These data and the clinical information associated with them will be provided to users by BBJ under certain conditions, after reviewing by the BBJ Sample and Data Access Committee as well as compliance with the security guidelines and contractual agreements.
| Data ID | JGAD000123 (JGAD000123) |
|---|---|
| Participants | BBJ 1st cohort: 182,505 patients |
| Targets | genome wide SNVs |
| Platform | Illumina [HumanOmniExpress、HumanExome、OmniExpressExome BeadChip] |
| Data ID | JGAD000529 (JGAD000529) |
|---|---|
| Participants | 1st cohort: 11,716 patients (47 diseases) 2nd cohort: 42,689 patients (38 diseases) |
| Targets | genome wide SNPs |
| Platform | Illumina [Asian Screening Array (ASA-24v1-0_A2) |
| No. of marker | 657,060 SNVs(GRCh38) |
Regarding the application process for using the genomic and omics analysis data available at BBJ Database, please refer to the following pages for details.
Application Process for Obtaining Samples and Data from BBJ
Where to find the data:
NBDC human database
Genome group sharing Database (AGD)
AMED CANNDs (Japanese only)
❁AGD is a service for archiving and sharing of all types of individual-level genetic and de-identified phenotypic data within research project and group members. The data will be shared among researchers with an approval for access by Human Data Review Board of NBDC.
Details on cohort information is available at the webpage mentioned below.
Our History
Controlled-access data is available only for use for research purposes. The access-granted researchers must have research experience in related studies and can use the data for their studies, after clarifying information such as the purpose of data use and data users. For use, approval is required at the review by the NBDC Data Access Committee as well as compliance with the security guidelines.
NBDC Research ID: hum0014
| Data ID | Type of Data・Participants/Materials・Release date | Cohort |
|---|---|---|
| JGAS000101 (JGAD000101: Phenotype, JGAD000102: Genotype) | Genotype and phenotype data for 8180 AF patients (2017/5/18) | 1 |
| JGAS000114 (JGAD000123) | BMI data for 158,284 individuals / Genotype data for 182,505 individuals (2017/5/18) | 1 |
| JGAS000114 (JGAD000144 – 201) | 162,255 individuals for 58 quantitative traits (2018/5/1) | 1 |
| JGAS000114 (JGAD000220: fastq) JGAD000410: bam,vcf) | WGS for 1,026 individuals (2018/8/13) | 1 |
| JGAS000140 (JGAD000209) | target sequencing of 11 hereditary breast cancer genes in 7,104 breast cancer patients and 23,731 controls (2018/10/16) | 1,2 |
| JGAS000114 (JGAD000220) | a reference panel from WGS data of the biobank Japan project (N=1,037) and 1KGP p3v5 ALL (N=2,504) (2019/9/27) | 1 |
| JGAS000203 (JGAD000288) | target sequencing of 8 hereditary prostate cancer genes in 7,636 prostate cancer patients and 12,366 controls (2019/10/7) | 1,2 |
| JGAS000293 (JGAD000399: target capture) (JGAD000400: SNP array) | target sequencing of 23 genes related to clonal hematopoiesis and SNP array in 11,234 subjects extracted from approximately 200,000 subjects registered in BioBank Japan between fiscal years 2003 to 2007 (2021/5/21) | 1 |
| JGAS000114 (Data addition) (JGAD000410) | bam/gvcf data of WGS (JGAD000220) 1,026 individuals(2021/07/13) | |
| JGAS000327 (JGAD000438: pancreatic cancer) | target sequencing of 27 cancer-predisposing genes in 1,005 pancreatic cancer patients (2021/11/26) | 1,2 |
| JGAS000346 (JGAD000458: colorectal cancer) | target sequencing of 27 cancer-predisposing genes in 12,503 colorectal cancer patients and 23,705 controls(2021/11/26) | 1,2 |
| JGAS000381 (JGAD000495: fastq, JGAD000496: bam, vcf) | WGS for 1,765 myocardial infarction patients and 199 dementia patients (2022/1/25) | 1,2 |
| JGAS000414 (JGAD000531) | target sequencings of 27 cancer-predisposing genes and 13 renal cell carcinoma-related genes in 740 renal cell cancer patients and 5,996 controls (2022/4/1) | 1,2 |
| JGAS000347 (JGAD000460) | target sequencing of 27 cancer-predisposing genes in 1,982 lymphoma patients (2023/4/20) | 1,2 |
| JGAS000592 (JGAD000720) | target sequencing of 27 cancer-predisposing genes in 10,366 gastric cancer patients (2023/4/20) | 1,2 |
| JGAS000592 (JGAD000721) | target Capture Sequencingデータ(target sequencing of 27 cancer-predisposing genes in 37,592 controls (2023/4/20) | 1,2 |
| JGAS000647 (JGAD000777) | WGS for 1,007 individuals from BBJ 1st cohort (47 diseases) (2024/1/11) | 1 |
| JGAS000698 (JGAD000831) | 256 gastric cancer (ICD10: C16) patients registered in BBJ 1st cohort (2024/05/27) | 1 |
| JGAS000699(JGAD000832) | WGS for 617 colorectal cancer patients (2024/05/27) | 1,2 |
| JGAS000700(JGAD000833) | low-depth WGS for 2,162 diabetes patients (2024/5/27) | 1 |
| JGAS000701(JGAD000834) | low-depth WGS for 2,067 gastric cancer patients (2024/5/27) | 1 |
| JGAS000703(JGAD000836) | SNP array for 269,000 patients (51 diseases) in BBJ 1st and 2nd cohort (2024/5/27) | 1,2 |
| JGAD000867 hum0014.v34 | Processed data of JGAD000220 (reference panel) by JGA (data for the TogoImputation reference panel) (2024/9/19) | |
| JGAD000868 hum0014.v34 | Processed data of JGAD000495 (reference panel) by JGA (data for the TogoImputation reference panel) (2024/9/19) | |
| JGAS000738 (JGAD000873) | Imputation reference panel for 7,472 Japanese WGS and 2,504 1000 Genome Project data (2024/10/25) | 1 |
| JGAS000746(JGAD000881) | Imputation reference panel for 3,256 Japanese WGS and 2,504 1000 Genome Project data (2024/11/28) | 1 |
| JGAS000782 (JGAD000924) | target sequencings on the coding regions of TP53 in 140,597 individuals, including those with breast cancer, stomach cancer, colon cancer, heart failure, stroke, etc. (2025/5/9) |
NBDC Research ID: hum0238
| Data ID | Type of Data・Participants/Materials・Release date | Cohort |
|---|---|---|
| JGAS000240 (JGAD000340) | NGS (HHV-6 sequences in WGS 10 samples (2020/7/28) | 1 |
NBDC Research ID: hum0311.
| Data ID | Type of Data・Participants/Materials・Release date | Cohort |
|---|---|---|
| JGAS000412 (JGAD000529) | Genotype data for 11,716 patients from BBJ 1st cohort and 42,689 patients from BBJ 2nd cohort) (2021/11/30) | 1,2 |
| JGAS000541 (JGAD000660) | Imputation data and index data for 180,882 patients from BBJ 1st cohort (2022/07/28) | 1 |
| JGAS000561 (JGAD000683) | NMR metabolic biomarkers on serum samples from cumulative 43,830 patients (45,270 samples) : 1,285 patients (1,286 samples) + 2,570 patients (2,573 samples) + 40,907 patients (41,411 samples) from BBJ 1st cohort with 47 diseases (2023/04/24) | 1 |
| JGAS000785 (JGAD000927) | Proteome data for 3,050 BBJ participants from BBJ 1st cohort (2025/06/11) | 1 |
AGD Group Shared Data are data shared in the AMED Genome group shared Database (AGD) in a framework that allows sharing in academic research or research that contributes to the improvement of public health within a research project or research group at an earlier stage, prior to its release at NBDC. AGD group-shared data can only be applied for by researchers who have been engaged in related research and whose purpose of use is limited to academic research or research that contributes to the improvement of course generation as principal researchers. Compliance with security guidelines is required for use.
Group-Shared Data(AGD)
| Data ID | Type of Data・Participants/Materials・Release date | Cohort |
|---|---|---|
AGDS_000013 (AGDD_000017) | 心房細動(ICD10:I48):547症例(BBJ登録検体)NGS (WGS) (2023/01/18) | 1 |
NBDC Unrestricted-access Data are public data that can be used by anyone without any restrictions on access. No review is required for use, but the data must be limited to research use, personal identification is prohibited, and the use of the most recent data must be observed.
NBDC Research ID: hum0014.
| Data ID | Type of Data・Participants/Materials・Release date | Cohort |
|---|---|---|
| hum0014.v1.freq.v1 | (GWAS for MI) 1666 MI patients and 3198 controls (2014/9/30) | 1 |
| hum0014.v2.jsnp. 934ctrl.v1 | Genotype frequencies in 934 healthy individuals (JSNP data) (2015/12/28) | MRC |
| 35 diseases | Genotype frequencies in each disease (JSNP data) 35 diseases about 190 patients in each disease set (2015/12/28) | 1 |
| hum0014.v2.jsnp. 182ec.v1 | Genotype frequencies in 182 esophageal cancer patients (JSNP data) (2015/12/28) | 1 |
| hum0014.v2.jsnp. 92als.v1 | Genotype frequencies in 92 amyotrophic lateral sclerosis (ALS) patients (JSNP data) (2015/12/28) | 1 |
| hum0014.v3. T2DM-1.v1 | GWAS for T2DM [1]) 9817 T2DM patients and 6763 controls (2016/1/28) | |
| hum0014.v3. T2DM-2.v1 | (GWAS for T2DM [2]) 5,646 T2DM patients and 19,420 controls (2016/1/28) | |
| hum0014.v4.AD.v1 | (GWAS for AD) 1472 AD patients and 7966 controls (2016/2/2) | |
| hum0014.v5.AF.v1 | (GWAS for AF) 8180 atrial fibrillation patients and 28,612 controls (2017/5/18) | 1 |
| hum0014.v6.158k.v1 | (GWAS for BMI) 182,505 individuals (158,284 individuals for BMI study) (2017/9/8) | 1 |
| hum0014.v7.POAG.v1 | (GWAS for POAG) 3980 POAG patients (Male: 1,997, Female: 1,983) 18,815 controls (Male: 7,817, Female: 10,998) (2018/4/4) | 1 |
| hum0014.v8.58qt.v1 | (GWAS for 58 quantitative traits) 162,255 individuals for 58 quantitative traits (2018/5/1) | 1 |
| hum0014.v9.Men.v1 hum0014.v9.MP.v1 | (GWAS for age at menarche and menopause) 67,029 females with information on age at menarche 43,861 females with information on age at menopause (2018/8/7) | 1 |
| hum0014.v12. T2DMwN.v1 | (meta analysis of 2 GWASs for T2DM with diabetic nephropathy) [GWAS-1] 2,380 T2DM with diabetic nephropathy patients 5,234 T2DM without diabetic nephropathy patients [GWAS-2] 429 T2DM with diabetic nephropathy patients 358 T2DM without diabetic nephropathy patients (2018/12/10) | |
| hum0014.v13. T2DMmeta.v1 | meta analysis of 4 GWASs for T2DM) on 36,614 T2DM patients and 155,150 controls (2019/1/25) | |
| hum0014.v14. smok.v1 | (GWAS for smoking behaviour) 165,436 individuals whose smoking status is available (2019/3/26) | 1 |
| hum0014.v15.ht.v1 | GWAS (GWAS for height) 159,095 individuals (Male: 86,257, Female: 72,838) (2019/9/27) | 1 |
| hum0014.v17 | GWAS for 40 diseases (2019/10/8) | |
| hum0014.v18 | GWAS for Breast cancer (2019/11/26) | |
| hum0014.v19 | (GWAS for dietary habits) 165,084 individuals whose dietary habits status is available (13 dietary traits) (2020/4/20) | 1 |
| hum0014.v20.cad.v1 | GWAS for coronary artery disease (2020/8/17) | |
| hum0014.v21 | GWAS for coronary artery disease 25,892 coronary artery disease patients (ICD10: I20-25) and 142,336 controls (2020/8/25) | |
| hum0197.v3.gwas.v1 | Biobank Japan (n = 179,000), UK biobank (n = 361,000), FinnGen (n = 136,000), no. Phenotypes: 220(2021/3/22) | 1 |
| hum0197.v5.gwas.v1 | GWAS for 10 phenotypes 10形質のGWAS(初潮年齢、閉経年齢、アルブミン/グロブリン比、自己免疫疾患、推算糸球体濾過量、悪性腫瘍、非アルブミン蛋白、リン、喫煙本数、喫煙歴) (2021/12/21) | 1 |
| hum0197.v5.finemap.v1 | GWAS fine-mapping Biobank Japan (n = 179,000), no. Phenotypes: 79 (2021/12/21) | |
| hum0197.v10.gwas.v1 | Biobank Japan (n=161,801), UK biobank (n=377,583), no. Phenotypes: 9 Patients: Autoimmune [Rheumatoid arthritis (ICD10: M05), Graves’ disease (ICD10: C719), type I diabetes mellitus (ICD10: E10)] Allergy [asthma (ICD10: J45), Atopic dermatitis (ICD10: L20), Pollinosis (ICD10: J301)] Controls: non-autoimmune +non-allergy individuals (There is overlap among patients in each disease category))(2022/6/16) | |
| hum0014.v27.surv.v1 | GWAS for survival time in 137,693 individuals from BBJ 1st cohort (2022/12/31) | 1 |
| hum0014.v28.MEs.v1 | mobile element variations in 4,880 individuals from BBJ 1st cohort (2023/4/5) | 1 |
| hum0014.v29.AF.v1 | GWAS for 9,826 AF patients and 140,446 controls from BBJ 1st cohort GWAS meta-analysis for 77,690 AF patients and 1,167,040 controls from BBJ, European, FinnGen(2023/4/5) | |
| hum0197.v16.gwas.v1 | large-scale meta-analysis including the summary statistics of other cohorts (FinnGen, BCAC, and PRACTICAL) for breast and prostate cancer (n=648,746 and 482,080) no. Phenotypes: 15 Patients: biliary tract (ICD10: C22.1, 23-24), breast (ICD10: C509, cervical (ICD10: C53), colorectal (ICD10: C18-20), endometrial (ICD10: C54), esophageal (ICD10: C15), gastric (ICD10: C16), hepatocellular (ICD10: C22.0), lung (ICD10: C34), non-Hodgkin’s lymphoma (ICD10: C82-83), ovarian (ICD10: C56), pancreatic (ICD10: C25), and prostate (ICD10: C61) cancer Controls: without cancer individuals (There is overlap among patients in each disease category) (2023/6/6) | |
| hum0197.v19.prs.v1 | The weights of variants calculated from GWAS results on type 2 diabetes (2024/5/29) | |
| hum0197.v20.gwas.v1 | GWAS for recurrent pregnancy loss (2024/5/30) | |
| hum0197.v21.gwas-jomon.v1 | GWAS for autoimmune diseases (2024/10/28) | |
| hum0197.v23.gwas-nmosd.v1 | Summary statistics of the genome-wide meta-analysis of NMOSD (2024/12/18) |
Summary data from genome-wide association study (GWAS), mainly for Japanese population, is available from RIKEN.
PheWeb releases genome-wide association study (GWAS) summary statistics on the BioBank Japan Project (BBJ). GWAS results in the Japanese population (mainly from BBJ) using the PheWeb platform are provided, with public access to the full summary statistics.
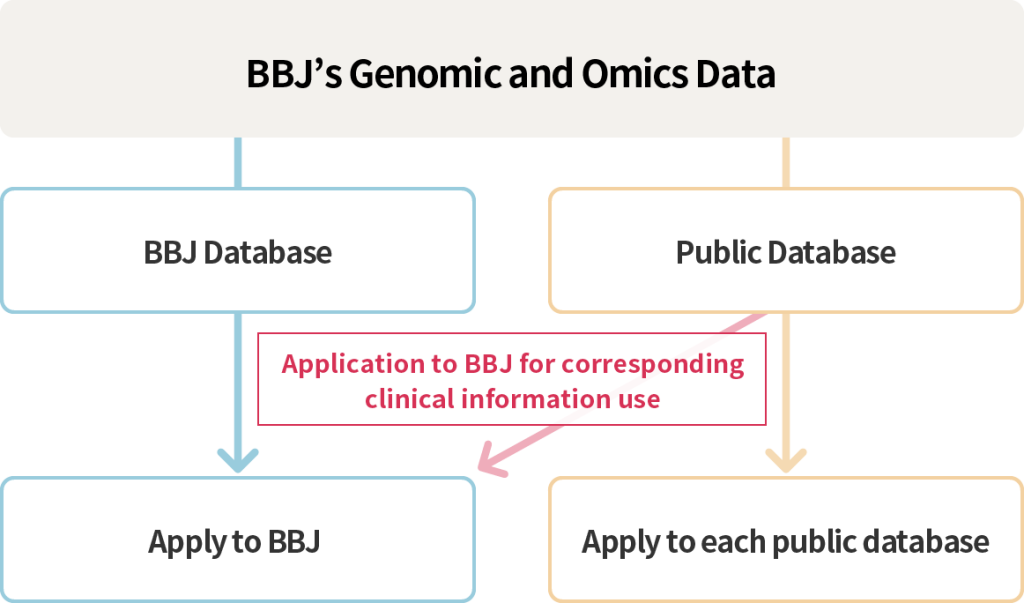
For data being available at BBJ database, please apply for use to BBJ. For data available at public databases, apply for use to the respective database.
For clinical information associated to the data you have applied to use from a public database, please follow the process mentioned at the Application Process for Obtaining Samples and Data from BBJ.
For detail of the clinical information, please refer to the following page (Japanese Only).
Details on the clinical information (basic information and disease sheet
Contact for inquiries concerning samples and data :
E-mail: shiryo_h “at “biobankjp.net
(Please change “at” to “@” to send e-mail.)